Visit Xiao Wang’s Google Scholar Page
Xiao also has a orcid page
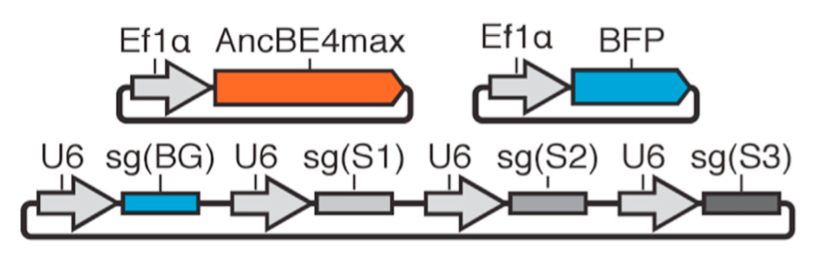 | Brookhouser N†, Tekel SJ†, Standage-Beier K†, Nguyen T, Schwarz G, Wang X, Brafman D. BIG-TREE: Base-Edited Isogenic hPSC Line Generation Using a Transient Reporter for Editing Enrichment. Stem Cell Reports 2020, 14, 184-191 | |
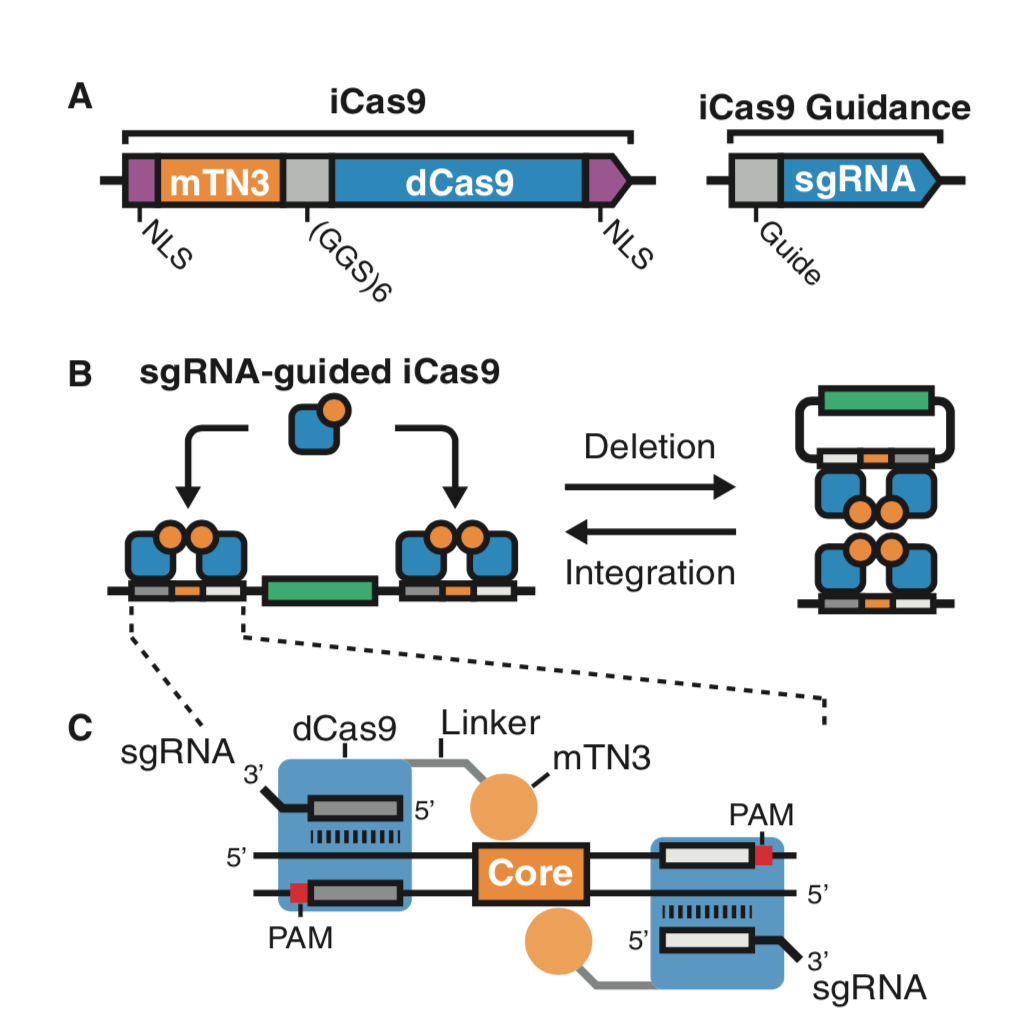 | Standage-Beier K, Brookhouser N, Balachandran P, Brafman D∗, and Wang X∗. RNA-Guided Recombinase-Cas9 Fusion Targets Genomic DNA Deletion and Integration. The CRISPR Journal 2019, 4, 209-222 | |
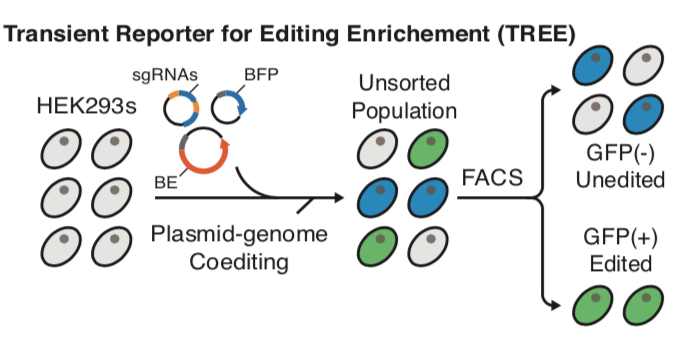 | Standage-Beier K†, Tekel SJ†, Brookhouser N†, Schwarz G, Nguyen T, Wang X∗, and Brafman D∗. A transient reporter for editing enrichment (TREE) in human cells. Nucleic Acids Research 2019, 10.1093/nar/gkz713 | |
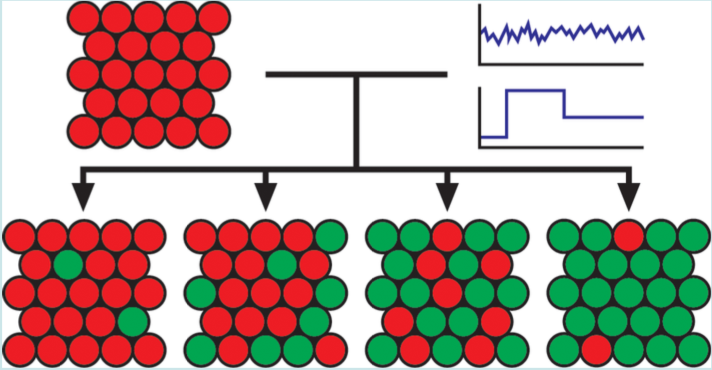 | Menn DJ, Sochor P, Goetz Hanah, Tian XJ∗, and Wang X∗. Intracellular Noise Level Determines Ratio Control Strategy Confined by Speed Accuracy Trade-off. ACS Synthetic Biology 2019, 6,1352-1360 | |
 | Menn DJ, Wang X. Modeling Gene Networks to Understand Multistability in Stem Cells. 2019, Computational Stem Cell Biology | |
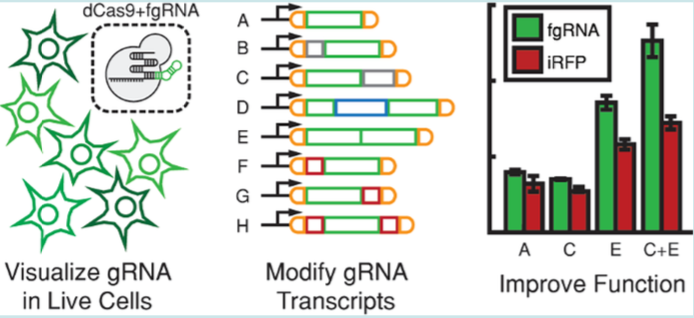 | Menn DJ, Pradhan S, Kiani S, Wang X. Fluorescent Guide RNAs Facilitate Development of Layered Pol II-Driven CRISPR Circuits. ACS Synthetic Biology 2018, 8, 1929-1936 | |
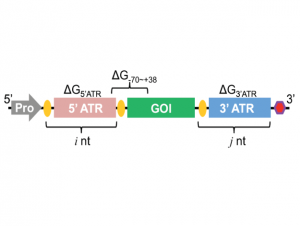 | Wu F, Zhang Q, Wang X. Design of Adjacent Transcriptional Regions to Tune Gene Expression and Facilitate Circuit Construction. Cell Systems 2018, 6(2), 206-215 | |
 | Menn DJ, Su R-Q, Wang X. Control of synthetic gene networks and its applications. Quantitative Biology. 2017, 5(2), 124-135, doi:10.1007/s40484-017-0106-5 | |
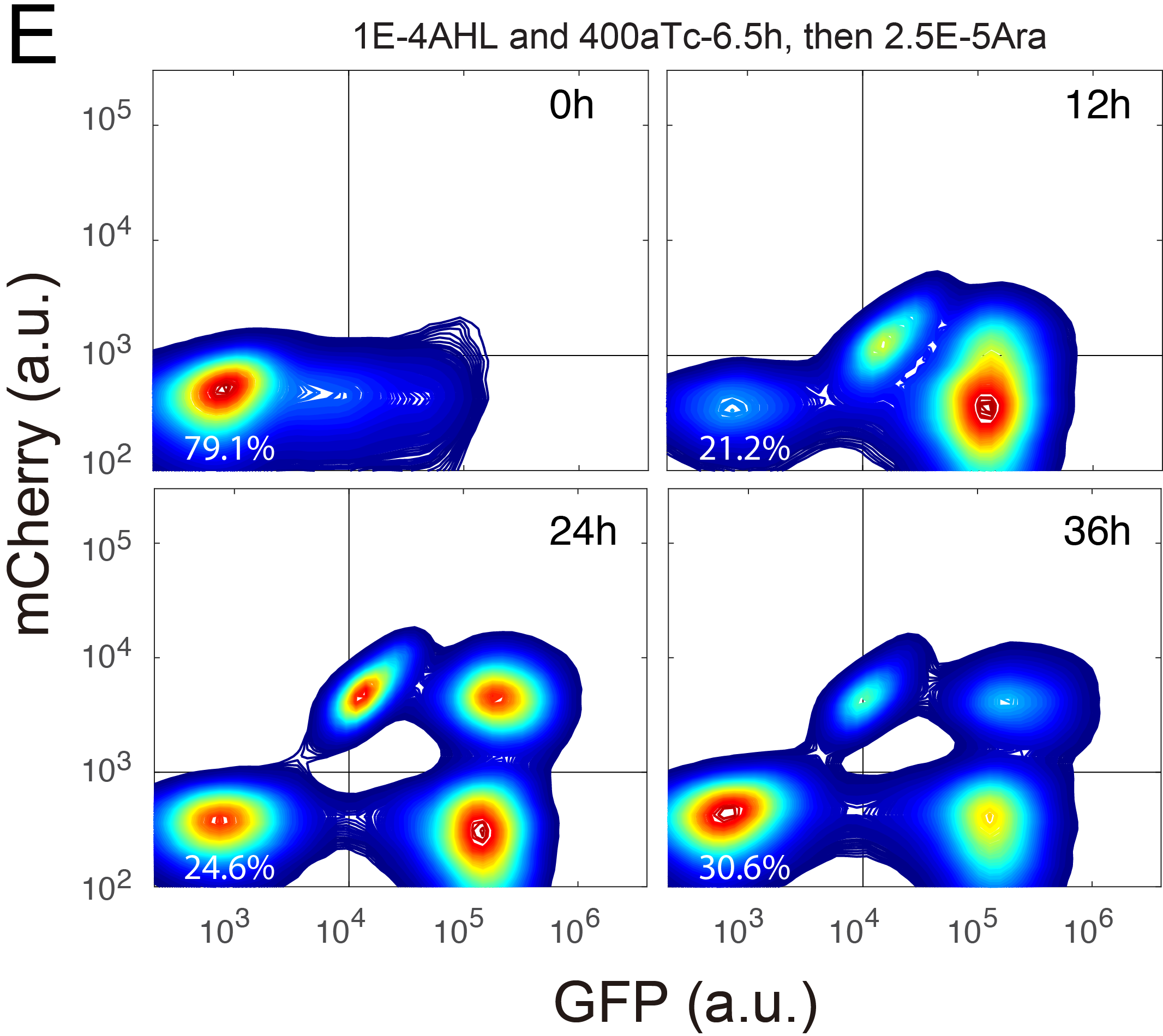 | Wu F, Su R-Q, Lai Y-C, Wang X. Engineering of a synthetic quadrastable gene network to approach Waddington landscape and cell fate determination. ELife. 2017, 6, e23702, doi:http://dx.doi.org/10.7554/eLife.23702 | |
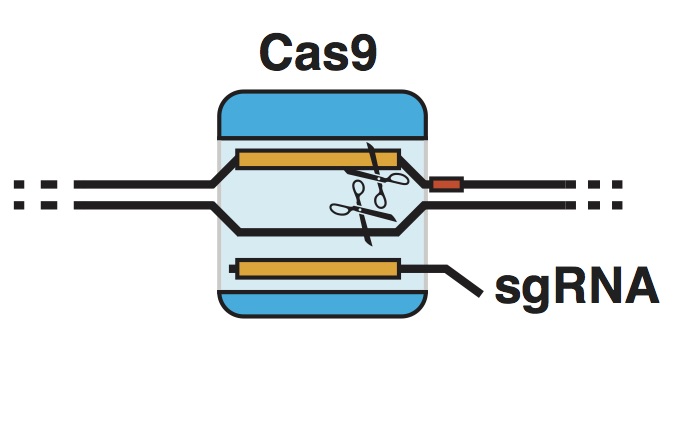 | Standage-Beier K, Wang X. Genome Reprogramming for Synthetic Biology. Frontiers of Chemical Science and Engineering. 2017, doi:10.1007/s11705-017-1618-2 | |
 | Wang L-Z, Su R-Q, Huang Z-G, Wang X, Wang W-X, Grebogi C and Lai Y-C. A geometrical approach to control and controllability of nonlinear dynamical networks. Nature Communications. 2016,7, doi:10.1038/ncomms11323 | |
 | Wang L-Z, Wu F, Flores K, Lai Y-C, Wang X. Build to understand: synthetic approaches to biology. Integrative Biology. 2016, 8, 394, doi: 10.1039/c5ib00252d | |
 | Standage-Beier K, Zhang Q, Wang X. Targeted Large-Scale Deletion of Bacterial Genomes Using CRISPR-Nickases. ACS Synthetic Biology. 2015, 4(11), 1217–1225 doi:10.1021/acssynbio. 5b00132 | |
 | Wu F, Wang X. Applications of synthetic gene networks. Science Progress,98(3), 244-252, doi:10.3184/003685015X14368807556 441 | |
 | Wu F, Menn DJ, Wang X. Quorum-Sensing Crosstalk-Driven Synthetic Circuits: From Unimodality to Trimodality. Chemistry & biology,21(12), 1629-1638 doi: 10.1016/j.chembiol.2014.10.008 (2014) | |
 | Faucon PC, Pardee K, Kumar RM, Li H, Loh Y-H, Wang X. Gene Networks of Fully Connected Triads with Complete Auto-Activation Enable Multistability and Stepwise Stochastic Transitions. PLoS ONE, 9(7), e102873. doi:10.1371/journal.pone.0102873 (2014) | |
 | Menn DJ, Wang X. Stochastic and Deterministic Decision in Cell Fate. eLS. (2014) | |
 | Su R-Q, Lai Y-C, Wang, X. Identifying Chaotic FitzHugh–Nagumo Neurons Using Compressive Sensing. Entropy, 16(7), 3889-3902. (2014) | |
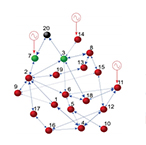 | Su R-Q, Lai Y-C, Wang X, and Do Y. Uncovering hidden nodes in complex networks in the presence of noise. Sci. Rep. 4. (2014) | |
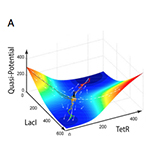 | Wu M, Su R-Q, Li X, Ellis T, Lai Y-C, Wang X. Engineering of regulated stochastic cell fate determination. Proceedings of the National Academy of Sciences USA 110:10610-10615 (2013) | |
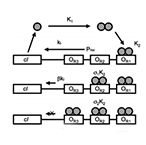 | Dari A, Kia B, Wang X, Bulsara A, Ditto W. Noise-aided computation within a synthetic gene network through morphable and robust logic gates. Physical Review E – Statistical, Nonlinear, and Soft Matter Physics 83(4) (2011) | |
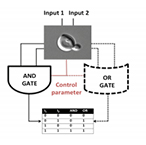 | Dari A, Bulsara A, Ditto W., Wang X. Reprogrammable biological logic gate that exploits noise. IEEE Biomedical Circuits and Systems Conference, BioCAS:337-340 (2011) | |
 | Murphy K, Adams R., Wang X, Balazsi G, Collins J. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Research 38(8):2712-2726 (2010) | |
 | Friedland AE*, Lu TK*, Wang X, Shi D, Church GM and Collins JJ. Synthetic Gene Networks That Count. Science 324:1199-1202 (2009) (* equally contributed) | |
 | Ellis T*, Wang X* and Collins JJ. Diversity-based, Model-guided Construction of Synthetic Gene Networks with Predicted Functions. Nature Biotechnology 27: 465-471 (2009). (* equally contributed) | |
 | Ellis T, Wang X and Collins JJ. Gene Regulation: Hacking the Network on a Sugar High. Molecular Cell 30:1-2 (2008) | |
 | Wang X, Errede B and Elston TC. Mathematical Analysis and Quantification of Fluorescent Proteins as Transcriptional Reporters. Biophysical Journal 94:2017-2026 (2008) | |
 | Guido NJ, Lee P, Wang X, Elston TC and Collins JJ. A Pathway and Genetic Factors Contributing to Elevated Gene Expression Noise in Stationary Phase. Biophysical Journal 93:L55-L57 (2007) | |
 | Erban R, Frewen T, Wang X, Elston TC, Coifman R, Nadler B and Kevrekidis I. Variable-free Exploration of Stochastic Models: A Gene Regulatory Network Example. Journal of Chemical Physics 126:155103 (2007) | |
 | Wang X, Hao N, Dohlman H and Elston TC. Bistability, Stochasticity, and Oscillations in the Mitogen-Activated Protein Kinase Cascade. Biophysical Journal 90:1961-1978 (2006) | |
 | Guido NJ*, Wang X*, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC and Collins JJ. A Bottom-Up Approach to Gene Regulation. Nature 439:856-860 (2006). (* equally contributed) |

